Gibson SOLA Platform
Make the DNA you want, when you want it
Synthetic Biology Meets AI to Craft Life with Computational Precision
Breaking the Bottleneck: On-Demand Biology in the Age of AI
Synthetic biology and machine learning might sound like buzzwords from science fiction, but together they’re reshaping how scientists create and modify organisms. Imagine being able to program bacteria to produce medicine, predict exactly how gene editing will turn out, or design entirely new biological entities. Thanks to artificial intelligence (AI), these futuristic scenarios are becoming today’s reality.
Metabolic Pathway Optimization
One key area significantly impacted by artificial intelligence (AI) in synthetic biology is metabolic pathway optimization. Metabolic pathways are the series of chemical reactions that occur inside cells, converting nutrients into energy, building blocks, or specific products like biofuels, pharmaceuticals, or industrial chemicals. Optimizing these pathways involves adjusting cellular metabolism to boost the production of desired compounds. Previously, this process required extensive experimentation using traditional methods of trial and error. It was necessary to systematically test different genetic modifications in cells such as bacteria or yeast, observing which alterations led to higher yields. Unfortunately, this conventional approach was slow, expensive, and inefficient.
Today, machine learning algorithms have transformed this scenario. These AI models are trained on massive datasets containing genomic data, experimental outcomes, and biochemical knowledge. With this information, they can precisely predict the impact of genetic changes on a cell’s metabolic functions. Instead of time-intensive trial and error, it is now possible to receive targeted guidance on which genes to modify for optimal results. This computational approach drastically reduces the experimental workload, accelerates development timelines, and cuts costs. Consequently, AI-driven metabolic pathway optimization empowers synthetic biologists to quickly and reliably engineer microbes that efficiently produce valuable substances, greatly enhancing both research capabilities and industrial applications.
Mapping Metabolic Pathways
Flux balance analysis (FBA) is a computational approach to analyze how substances flow within a cell’s metabolic networks. Think of it as constructing a comprehensive map illustrating the precise routes that molecules, like nutrients or metabolites, take inside cells such as bacteria or yeast. Each cell contains numerous interconnected pathways, each performing specific functions like generating energy, discarding waste, or synthesizing valuable products like pharmaceuticals (figure 1). Historically, enhancing these pathways to boost production involved extensive trial-and-error experimentation. It was necessary to manually modify genes without certainty of success, resulting in a lengthy, costly, and uncertain process.
Recently, scientists have begun integrating FBA with machine learning algorithms to significantly streamline this process. Machine learning systems can analyze vast amounts of biological data, including genetic sequences and outcomes from prior experiments, enabling precise predictions about how specific genetic changes will influence metabolic outputs. This advanced approach eliminates much of the uncertainty involved, providing scientists with accurate, evidence-based guidance on the best genetic modifications to implement. Consequently, this powerful combination of AI and FBA has dramatically accelerated scientific discovery, enabling more efficient microbial engineering for producing medications, renewable energy sources, and various industrially relevant compounds, while simultaneously lowering costs and improving accessibility.
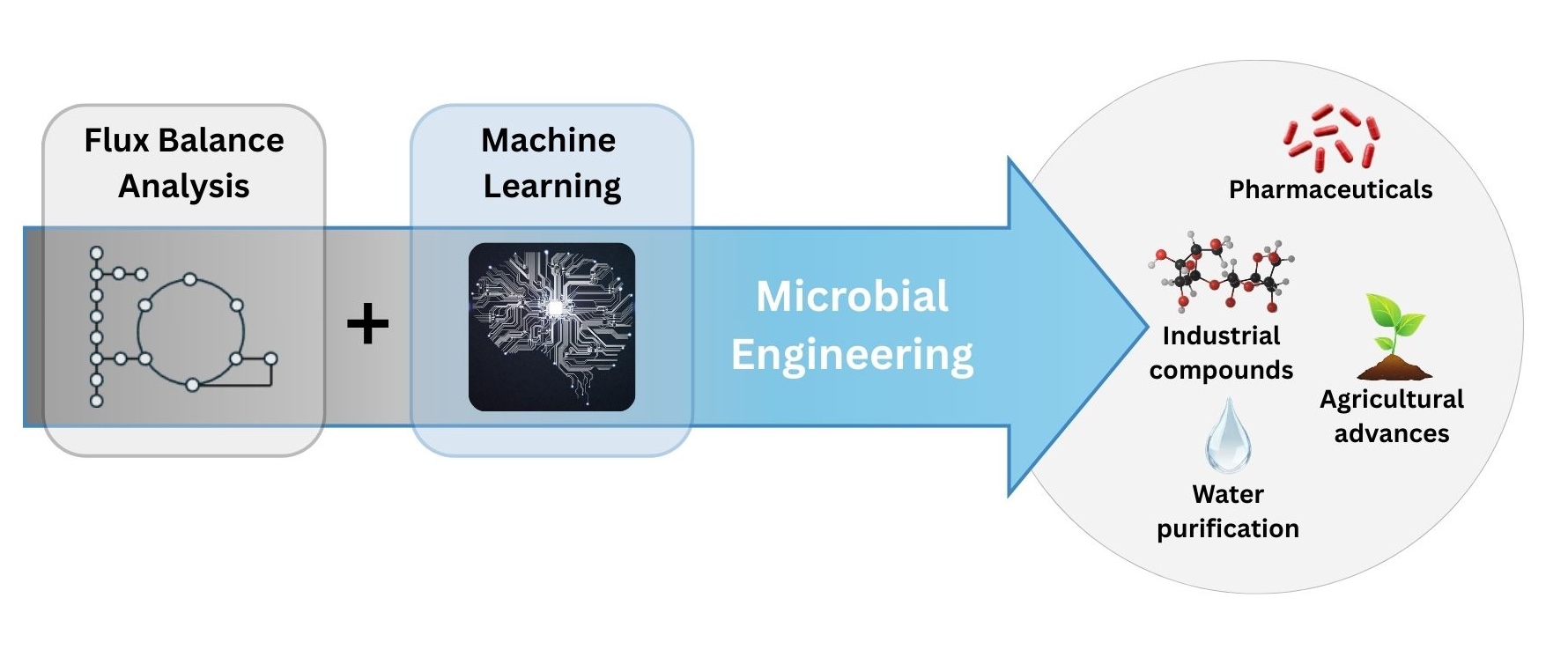
Figure 1. Computational Flux Balance Analysis. AI models accelerate pathway determination for microbial engineering in applications ranging from drug development to industrial compound engineering.
Streamlining Antibody Discovery
Similar to how scientists focused on Metabolomics are using AI to shift from serendipitous discovery toward rational, data-driven discovery, AI and ML are revolutionizing traditional antibody discovery workflows. By dramatically accelerating the design, screening, and optimization of therapeutic antibodies scientists can drive towards more efficient use of resources and improve overall productivity. Traditionally, antibody discovery relied on time-consuming processes such as hybridoma screening or phage display, which required labor-intensive wet-lab work and iterative trial-and-error optimization. AI now enables in silico prediction of antigen–antibody binding, immunogenicity, and developability, allowing researchers to narrow down candidate pools with far greater efficiency and precision before even entering the lab.
These models trained on massive datasets such as antibody sequence–structure–function relationships that can identify patterns and predict high-affinity, specific binders and also support virtual screening and affinity maturation, dramatically reducing the time and cost needed to advance a lead candidate (figure 2). These tools are ultimately streamlining development pipelines and enabling more personalized and potent therapies while getting new therapies to market faster and with reduced developmental cost.

Figure 2. AI Models Predict Higher Probability Antibody Targets. AI models can identify patterns and predict high-affinity, specific binders and also support virtual screening and affinity maturation.
Gene Editing
In addition to streamlining traditional workflows, AI is significantly improving the safety and accuracy of genome editing, particularly when combined with CRISPR technology. While CRISPR revolutionized biological research by enabling precise genetic modifications, it still faces significant challenges—particularly unintended edits or off-target effects. These accidental alterations can lead to unwanted side effects that can be potentially harmful in therapeutic settings or problematic in genetically modified crops.
AI, specifically machine learning, is addressing this challenge head-on. Machine learning models analyze extensive datasets containing genetic sequences and past editing outcomes to identify patterns that human researchers might miss. By detecting subtle relationships between gene sequences and editing accuracy, these algorithms predict where unintended edits are likely to occur. With these insights, it’s now possible to carefully select CRISPR targets and design experiments that dramatically reduce the chances of errors.
The practical implications of this AI-driven precision are enormous. In medicine, it means gene therapies can become safer, paving the way for treatments of genetic disorders without the fear of harmful side effects. In agriculture, genetically modified crops can be developed with greater confidence, enhancing their nutritional value or resilience to climate conditions. Ultimately, by making genome editing safer and more reliable, AI amplifies CRISPR’s potential to beneficially transform human health and agriculture.
Removing Barriers and Delays
One exciting innovation breaking barriers in synthetic biology is overnight DNA synthesis using the Gibson SOLA Platform by Telesis Bio. Traditionally, synthesizing precise DNA sequences required substantial time, delaying research and limiting experimentation flexibility. However, the SOLA Platform dramatically changes this by enabling rapid, automated DNA synthesis directly within researchers’ labs overnight. This technology combines state-of-the-art enzymatic methods with advanced robotics, removing the dependency on external service providers and shipping delays (figure 3).
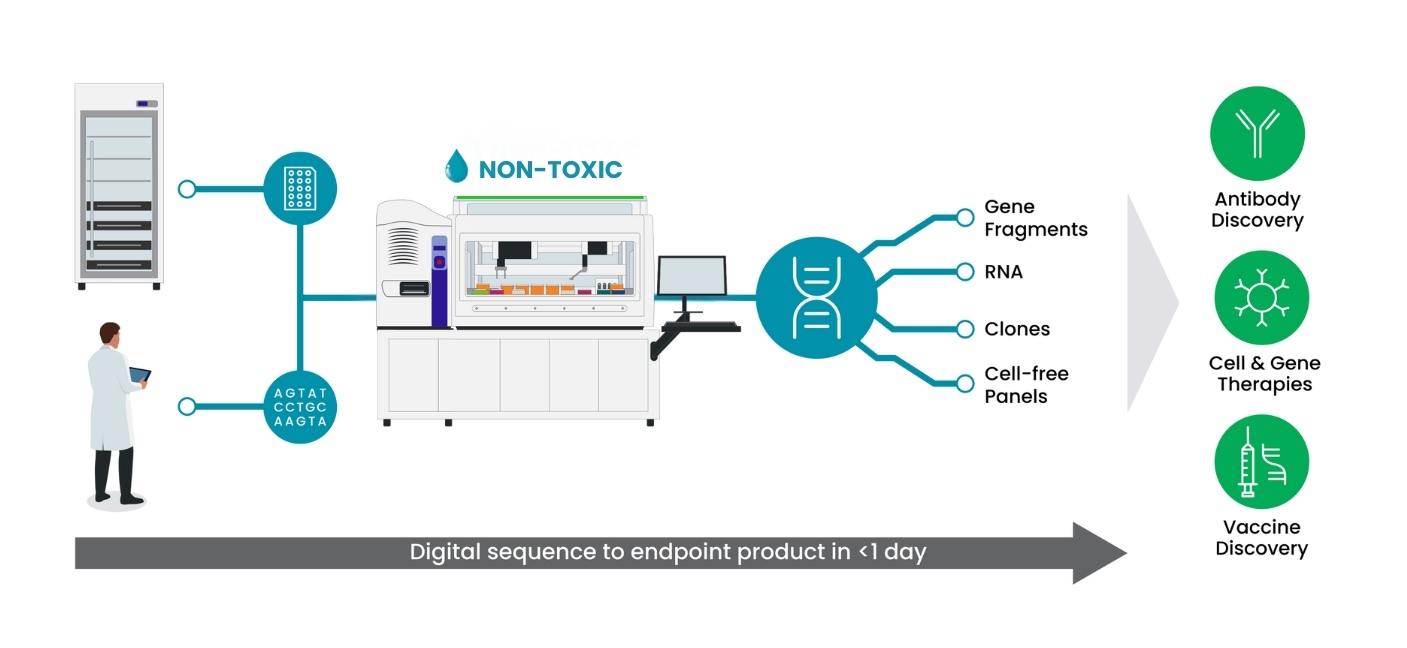
Figure 3. Digital Sequence to Biological Endpoint in 1 Day. The Gibson SOLA Platform is a ‘digital to biological converter’ that generates DNA or mRNA in less than one day.
The Gibson SOLA Platform streamlines workflows significantly, making it possible to quickly design, test, and iterate genetic constructs within just one day. This acceleration allows researchers to rapidly advance their therapeutic development and protein engineering projects, speeding up the overall pace of scientific discovery. Furthermore, it reduces logistical hurdles like supply chain disruptions or delays caused by external providers. When paired with automation tools like the Echo 525 Acoustic Liquid Handler and the Biomek i7 Automated Workstation from Beckman Coulter Life Sciences, labs can consistently output more than 50kb of DNA per week.
By making DNA synthesis fast, reliable, and immediately accessible, Telesis Bio’s Gibson SOLA Platform not only enhances research productivity but also lowers barriers to innovation, allowing labs to participate actively in groundbreaking synthetic biology research.
For more information on how the Gibson SOLA Platform is transforming DNA synthesis, visit telesisbio.com/sola.
What could you build with a process that is >93% faster?
Determine how Gibson SOLA could empower your team with a personalized 1:1 consultation
Meet with our experts to determine how Gibson SOLA could help you to build and test more quickly.

1:1 Consultations available to determine how Gibson SOLA could enable your team.